You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
The A.I Megathread (LLM , GPT , Development)
More options
Who Replied?
Adobe expands Firefly with new Generative Fill feature
Generative AI tools can be supportive, helping a wide range of people reach their goals in various creative efforts. Adobe certainly hopes that is the case, as it introduces a brand new tool to bolster its artificial intelligence efforts.
Adobe launches Generative Fill to boost generative AI image generation
| May 23, 2023An image generated by Generative Fill

Generative AI tools can be supportive, helping a wide range of people reach their goals in various creative efforts. Adobe certainly hopes that is the case, as it introduces a brand new tool to bolster its artificial intelligence efforts.
Adobe introduced Firefly, the family of generative AI models, earlier this year. With this tool, artists can simply input text to describe what they want to add to their creation. Want to put a deer inside an alleyway at night? Firefly can help you with that, with this new addition.
Today, Adobe is introducing Generative Fill directly into the Photoshop app, for beta users. The company says the feature will rollout more broadly in the second half of 2023.
As for the new Generative Fill feature, Adobe says it's designed to work as a co-pilot for users to streamline their creativity. Generative Fill allows users to non-destructively add, extend, or remove content from images as they're working. Just using simple text prompts, like "yellow road lines" can bring them into your image of a bicyclist.
Generative Fill works with each individual work. It automatically matches the style of the image the user is working on, while also matching lighting and perspective. Here are the major bullet points:"By integrating Firefly directly into workflows as a creative co-pilot, Adobe is accelerating ideation, exploration and production for all of our customers," said Ashley Still, senior vice president, Digital Media at Adobe. "Generative Fill combines the speed and ease of generative AI with the power and precision of Photoshop, empowering customers to bring their visions to life at the speed of their imaginations."
- Powered by Firefly: Firefly is designed to generate images that are safe for commercial use and is trained on Adobe Stock's hundreds of millions of professional-grade, licensed, high-resolution images, helping ensure Firefly won't generate content based on other peoples' or brands' intellectual property (IP).
- Magically leap from idea to image, with simple text prompts: Add, extend or remove content from images to achieve astounding results.
- Edit non-destructively: Create newly generated content in generative layers, enabling you to rapidly iterate through a myriad of creative possibilities and reverse the effects when you want, without impacting your original image.
- Create at a transformative rate: Experiment with off-the-wall ideas, ideate different concepts and produce boundless variations of high-quality content as fast as you can type.
- Available as a web tool: Generative Fill is also available as a new module in the Firefly beta for users interested in testing the new capabilities on the web.

New superbug-killing antibiotic discovered using AI
The drug can target one of the three most dangerous bacterial superbugs, say researchers.
www.bbc.com
New superbug-killing antibiotic discovered using AI
- Published
6 hours ago
- Published

Image caption,
Scientist Denise Catacutan working on the experimental antibiotic discovered with the help of artificial intelligence.
By James Gallagher
Health and science correspondent
Scientists have used artificial intelligence (AI) to discover a new antibiotic that can kill a deadly species of superbug.
The AI helped narrow down thousands of potential chemicals to a handful that could be tested in the laboratory.
The result was a potent, experimental antibiotic called abaucin, which will need further tests before being used.
The researchers in Canada and the US say AI has the power to massively accelerate the discovery of new drugs.
It is the latest example of how the tools of artificial intelligence can be a revolutionary force in science and medicine.
Stopping the superbugs
Antibiotics kill bacteria. However, there has been a lack of new drugs for decades and bacteria are becoming harder to treat, as they evolve resistance to the ones we have.
More than a million people a year are estimated to die from infections that resist treatment with antibiotics.
The researchers focused on one of the most problematic species of bacteria - Acinetobacter baumannii, which can infect wounds and cause pneumonia.
You may not have heard of it, but it is one of the three superbugs the World Health Organization has identified as a "critical" threat.
It is often able to shrug off multiple antibiotics and is a problem in hospitals and care homes, where it can survive on surfaces and medical equipment.
Dr Jonathan Stokes, from McMaster University, describes the bug as "public enemy number one" as it's "really common" to find cases where it is "resistant to nearly every antibiotic".

Image caption,
Dr Jonathan Stokes
Artificial intelligence
To find a new antibiotic, the researchers first had to train the AI. They took thousands of drugs where the precise chemical structure was known, and manually tested them on Acinetobacter baumannii to see which could slow it down or kill it.
This information was fed into the AI so it could learn the chemical features of drugs that could attack the problematic bacterium.
The AI was then unleashed on a list of 6,680 compounds whose effectiveness was unknown. The results - published in Nature Chemical Biology - showed it took the AI an hour and a half to produce a shortlist.
The researchers tested 240 in the laboratory, and found nine potential antibiotics. One of them was the incredibly potent antibiotic abaucin.
Laboratory experiments showed it could treat infected wounds in mice and was able to kill A. baumannii samples from patients.
However, Dr Stokes told me: "This is when the work starts."
The next step is to perfect the drug in the laboratory and then perform clinical trials. He expects the first AI antibiotics could take until 2030 until they are available to be prescribed.
Curiously, this experimental antibiotic had no effect on other species of bacteria, and works only on A. baumannii.
Many antibiotics kill bacteria indiscriminately. The researchers believe the precision of abaucin will make it harder for drug-resistance to emerge, and could lead to fewer side-effects.
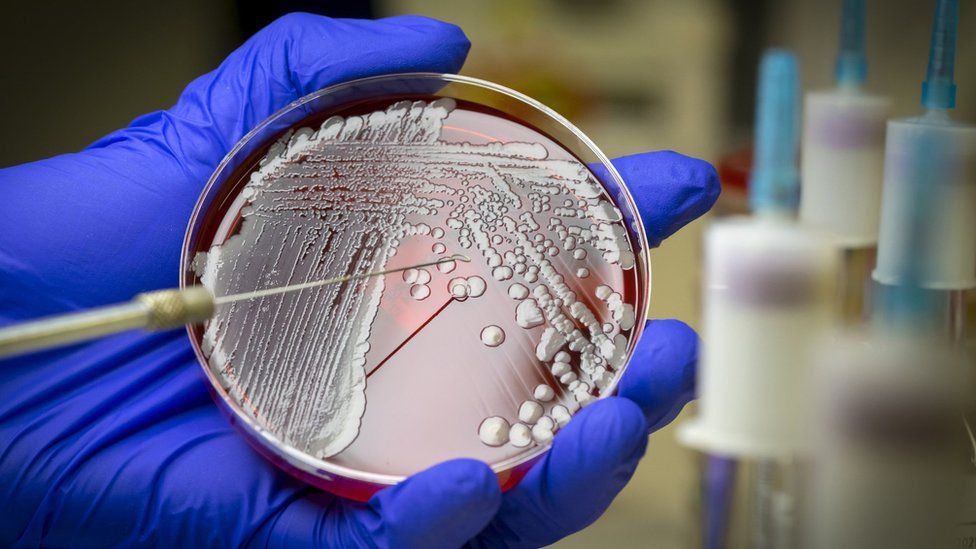
Bacteria being grown in the laboratory
In principle, the AI could screen tens of millions of potential compounds - something that would be impractical to do manually.
"AI enhances the rate, and in a perfect world decreases the cost, with which we can discover these new classes of antibiotic that we desperately need," Dr Stokes told me.
The researchers tested the principles of AI-aided antibiotic discovery in E. coli in 2020, but have now used that knowledge to focus on the big nasties. They plan to look at Staphylococcus aureus and Pseudomonas aeruginosa next.
"This finding further supports the premise that AI can significantly accelerate and expand our search for novel antibiotics," said Prof James Collins, from the Massachusetts Institute of Technology.
He added: "I'm excited that this work shows that we can use AI to help combat problematic pathogens such as A. baumannii."
Follow James on Twitter.
Multi-task Language Understanding on MMLU

Papers with Code - MMLU Benchmark (Multi-task Language Understanding)
The current state-of-the-art on MMLU is GPT-4 (few-shot, k=5). See a full comparison of 74 papers with code.
paperswithcode.com

AI + IQ testing (human vs AI)
Viz Download source (PDF) Tests: View the data (Google sheets) Trivia test (by WCT): Human: 52%, GPT-3: 73%, J1: 55.4% Notable events in IQ testing AI models Date Summary Notes May/2023 PaLM 2 breaks 90% on WinoGrande (PDF, Google) For the first time, a large language model has breached the 90%...

SuperGLUE Benchmark
SuperGLUE is a new benchmark styled after original GLUE benchmark with a set of more difficult language understanding tasks, improved resources, and a new public leaderboard.
SuperGLUE: A Stickier Benchmark for General-Purpose Language Understanding Systems

Researchers use AI models to advance drug delivery system for chronic eye diseases
Promising prospects for developing improved and more tolerable drug treatments for prevalent chronic eye diseases
 www.somaeu.com
www.somaeu.com
Researchers use AI models to advance drug delivery system for chronic eye diseases
Researchers from the Wilmer Eye Institute at Johns Hopkins Medicine have successfully utilized artificial intelligence models and machine-learning algorithms to predict the optimal amino acid components of therapeutic proteins for safe drug delivery to animal eye cells. This collaborative project with the University of Maryland offers promising prospects for developing improved and more tolerable drug treatments for prevalent chronic eye diseases like glaucoma and macular degeneration, affecting millions of individuals in the United States. The current drug therapies for these conditions, involving frequent eye injections or multiple daily eye drops, can be challenging to sustain and tolerate over time. Therefore, the scientific community has been focusing on developing delivery systems that can bind to eye cell components, extending the therapeutic effects of medications without compromising safety.
In 2020, the Food and Drug Administration approved an implantable device designed to release drugs for glaucoma treatment. Although this device offered longer-lasting effects compared to drops or injections, prolonged use in some cases led to eye cell death, necessitating a return to traditional methods like eye drops and injections.
The findings of the recent research, published in Nature Communications on May 2, demonstrate the accurate predictions made by AI-designed models regarding the ideal sequence of amino acids (peptides or small proteins) that can bind to specific chemicals in rabbit eye cells. This binding enables the safe and sustained release of medications over several weeks, reducing the need for strict and frequent treatment schedules. The focus of the investigation was specifically on peptides that bind to melanin, a pigment present in various specialized structures within eye cells.
The research team acknowledged previous studies on peptide-based drug delivery systems and their effectiveness. However, they aimed to identify peptides that could strongly bind to a widely distributed compound in the eye. To achieve this, they employed rapid machine learning techniques and artificial intelligence methods. Dr. Laura Ensign, the co-corresponding author of the paper and the Marcella E. Woll Professor of Ophthalmology at the Johns Hopkins University School of Medicine, explained that the team utilized machine learning algorithms to predict promising peptide sequences for drug delivery involving melanin.
The process began by inputting thousands of data points, including amino acid characteristics and peptide sequences, into the machine learning model. This allowed the model to "learn" the chemical and binding properties of different combinations of amino acids and eventually predict potential peptide sequences for effective drug delivery with melanin.
Through this artificial intelligence model, the researchers generated 127 peptides with varying abilities to penetrate specialized cells containing melanin, bind to melanin, and exhibit non-toxic properties. Among these peptides, the model identified HR97 as having the highest success rate in binding. The team also validated the properties of these peptides, such as their improved uptake and binding within cells, as well as the absence of any signs of cell death.
To test the model's prediction, the researchers attached HR97 to the drug brimonidine, which is used to treat glaucoma by reducing inner eye pressure, and administered it to adult rabbit eyes through injection. To assess the performance of HR97, the researchers measured the concentration of brimonidine in the eye cells by analyzing the drug's levels after administering the experimental drug delivery system. They discovered that significant amounts of brimonidine were present for up to one month, indicating that HR97 effectively penetrated the cells, bound to melanin, and released the drug over an extended period. Additionally, the team confirmed that when brimonidine was bound to HR97, it sustained its ability to lower eye pressure for up to 18 days, and there were no indications of eye irritation in the rabbits.
Dr. Ensign emphasized that future studies utilizing artificial intelligence to predict peptides for drug delivery could have significant implications for other conditions involving melanin and could be extended to target other specialized structures.
''We believe we are well on the way to finding solutions in trying to improve patient care and quality of life using drug delivery systems. The ultimate goal is creating something that we can translate out of the lab and actually make people's lives better."
Moving forward, according to Ensign, researchers will need to explore methods to further prolong the duration of drug action. They also aim to assess the success rate of the AI model's drug delivery predictions with different medications and evaluate safety in human subjects.
The study involved several other researchers, including Henry Hsueh, Usha Rai, Wathsala Liyanage, Yoo Chun Kim, Matthew Appell, Jahnavi Pejavar, Kirby Leo, Charlotte Davison, Patricia Kolodziejski, Ann Mozzer, HyeYoung Kwon, Maanasa Sista, Sri Vishnu Kiran Rompicharla, Malia Edwards, Ian Pitha, and Justin Hanes from the Johns Hopkins University School of Medicine. Additionally, Nicole Anders and Avelina Hemingway from the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center, as well as Renee Ti Chou and Michael Cummings from the University of Maryland, were involved in the research.

